Chapter 11 Expression in Specific pathways
The above plots suggest that there could be a different in growing and stable based on:
- inflammation
- Kras signalling
- MHC presentation
- checkpoint proteins
We can pull out the genes in these sets and visualise the relative expression in a heatmap in the DN, CD45 and Ep samples:
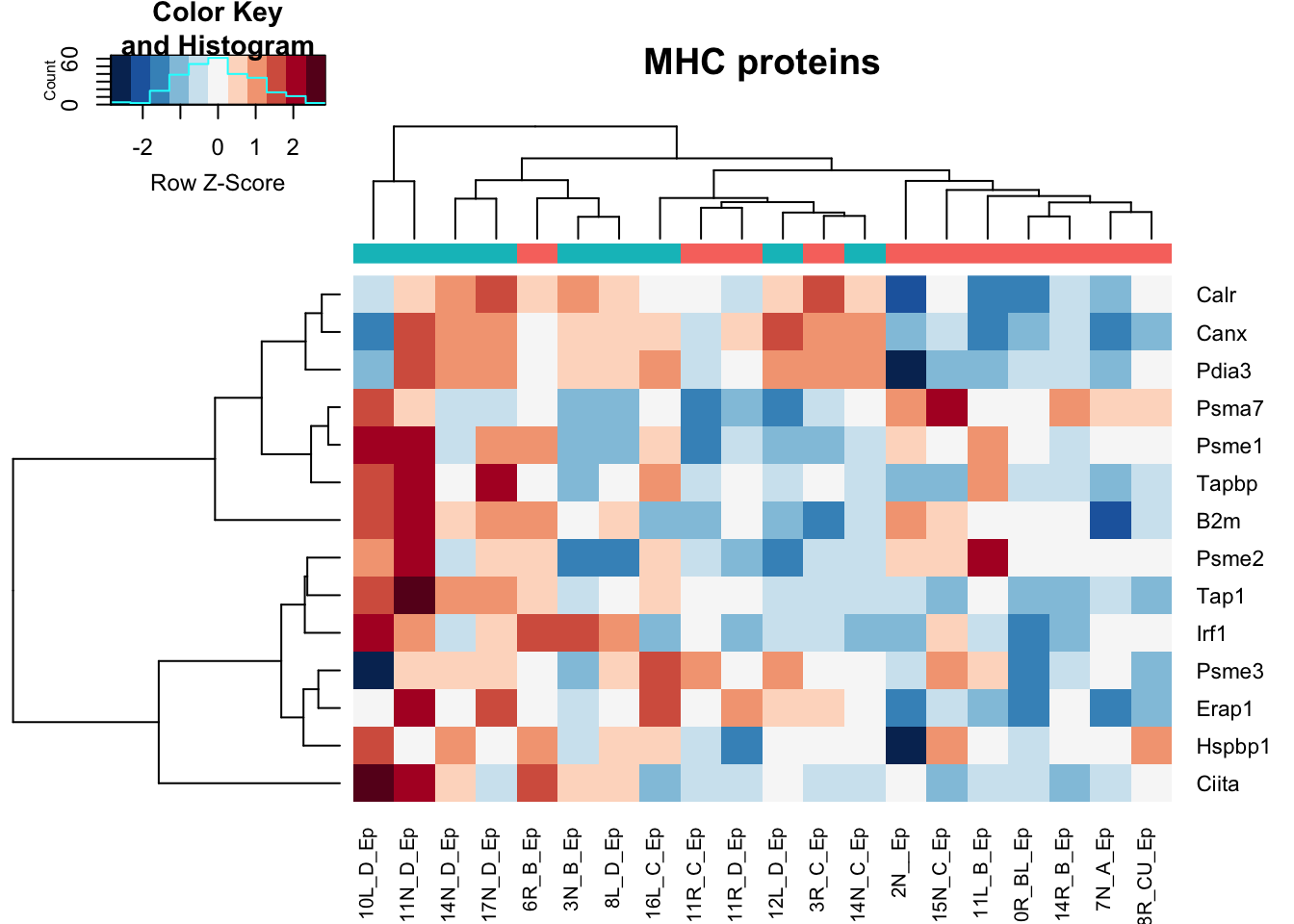
Note that red is growing and green is stable
# library(org.Hs.eg.db)
load("../anntotations/ListofGeneSets.RData")
SetNamesc2=names(PathInc2) #names(ListGSC$c2List)
x1=grep("MHC", SetNamesc2)
GeneNames=unique(unlist(geneIds(PathInc2[x1[c(1, 4,5)]])))
#test1=mapIds(org.Hs.eg.db, GeneNames, 'SYMBOL','ENTREZID')
RatGeneNamesMHC=na.omit(unique(SymHum2Rat$RGD.symbol[match(GeneNames, SymHum2Rat$HGNC.symbol)]))
#RatGeneNamesKras=mapIds(org.Hs.eg.db, ListGSC$Hallmark$HALLMARK_KRAS_SIGNALING_UP, 'SYMBOL','ENTREZID')
GeneNames=unlist(geneIds(PathInH["HALLMARK_KRAS_SIGNALING_UP"]))
RatGeneNamesKras=na.omit(unique(SymHum2Rat$RGD.symbol[match(GeneNames, SymHum2Rat$HGNC.symbol)]))
GeneNames=unlist(geneIds(PathInH["HALLMARK_INFLAMMATORY_RESPONSE"]))
RatGeneNamesInf=na.omit(unique(SymHum2Rat$RGD.symbol[match(GeneNames, SymHum2Rat$HGNC.symbol)]))
CheckpointProt=unlist(ImmSuppAPC)
RatGeneNamesCheckpoint=na.omit(unique(SymHum2Rat$RGD.symbol[match(CheckpointProt, SymHum2Rat$HGNC.symbol)]))11.0.0.1 DN samples:
Note the distribution of growing/stable in DN is: 21, 9
ColsideColsC=hue_pal()(2)[DNdds$Growth]
heatmap.2(assay(vstDN)[na.omit(match(RatGeneNamesMHC, rownames(vstDN))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC expression")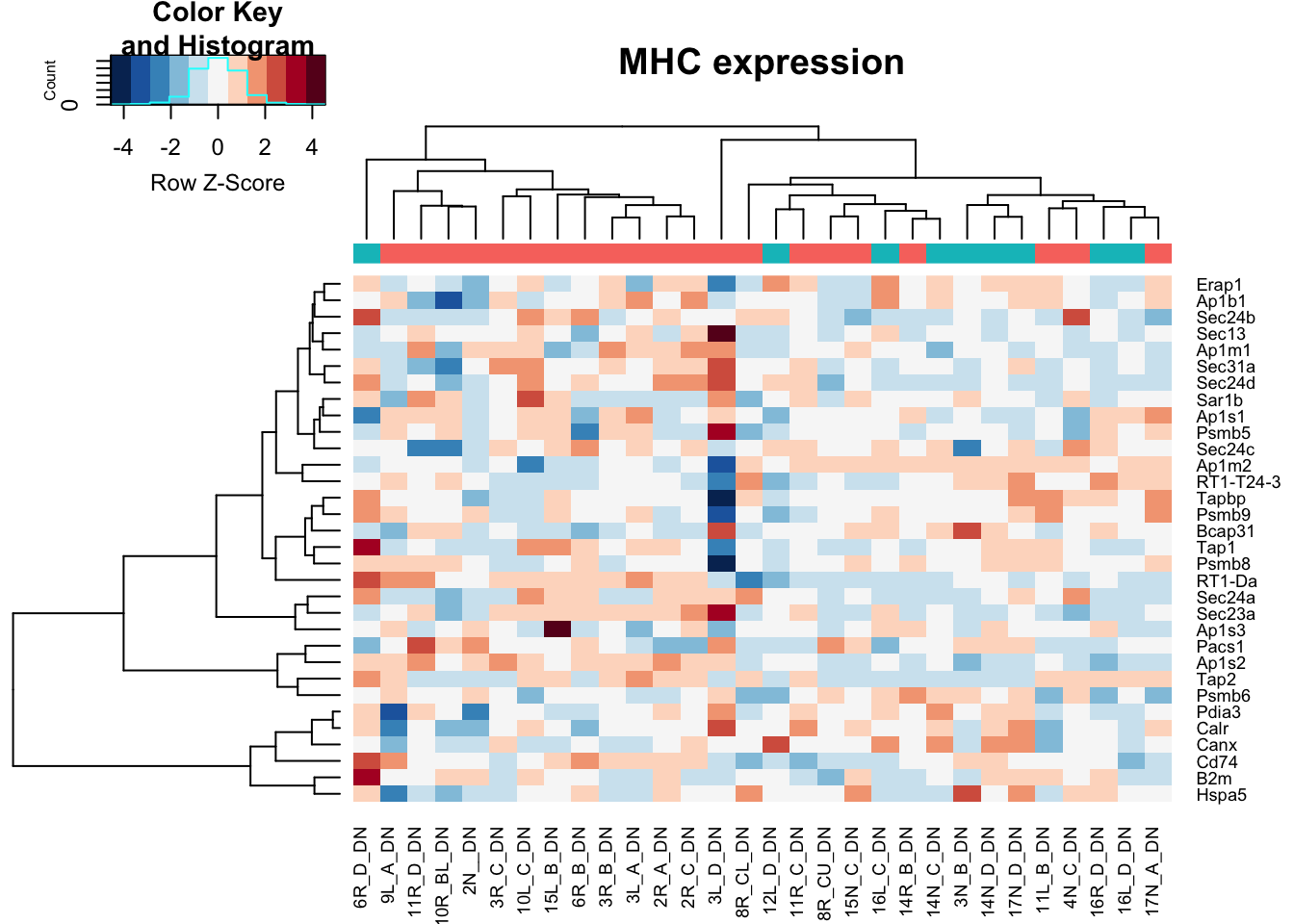
heatmap.2(assay(vstDN)[na.omit(match(RatGeneNamesKras, rownames(vstDN))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Kras expression")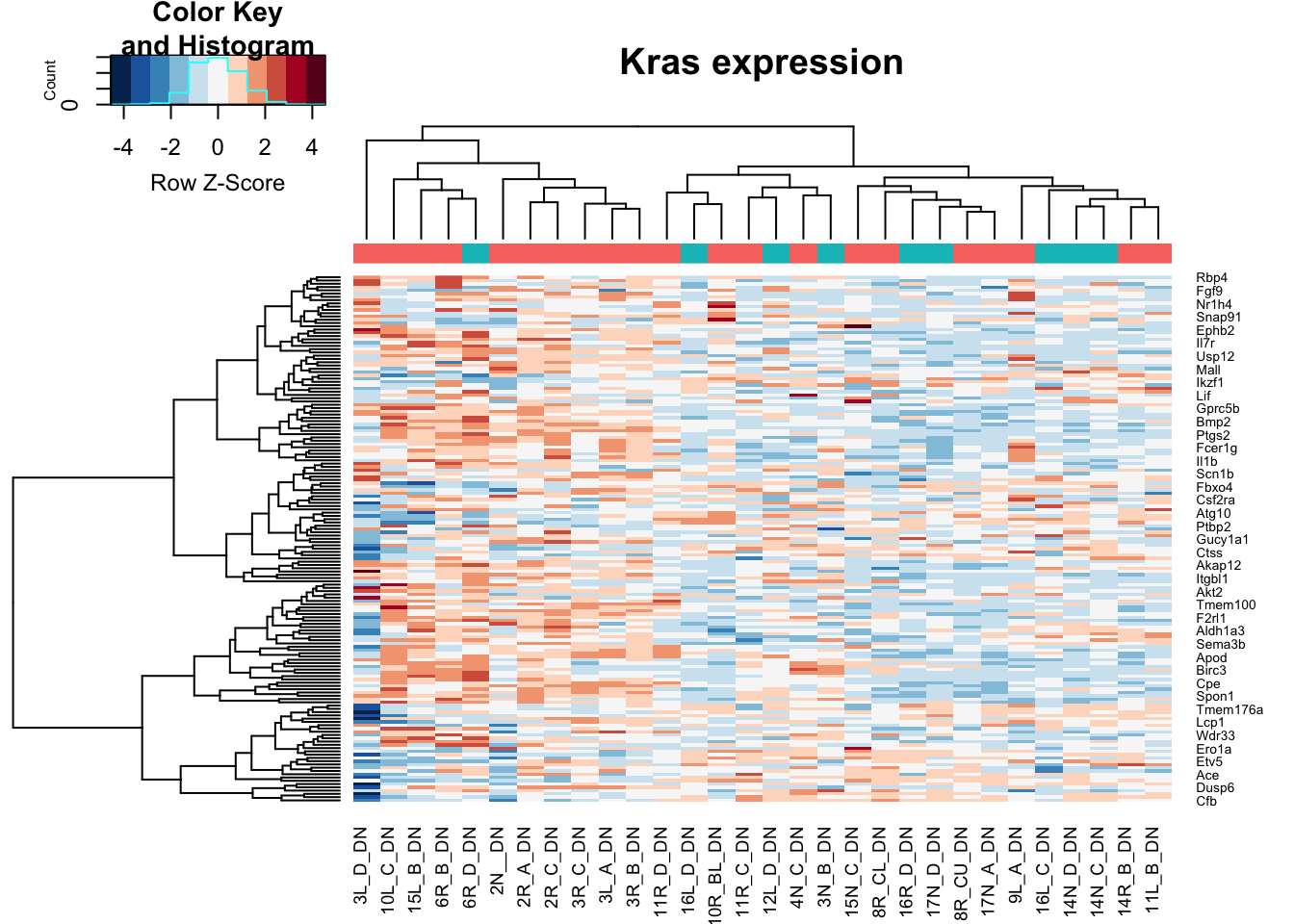
heatmap.2(assay(vstDN)[na.omit(match(RatGeneNamesInf, rownames(vstDN))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Inflammatory response")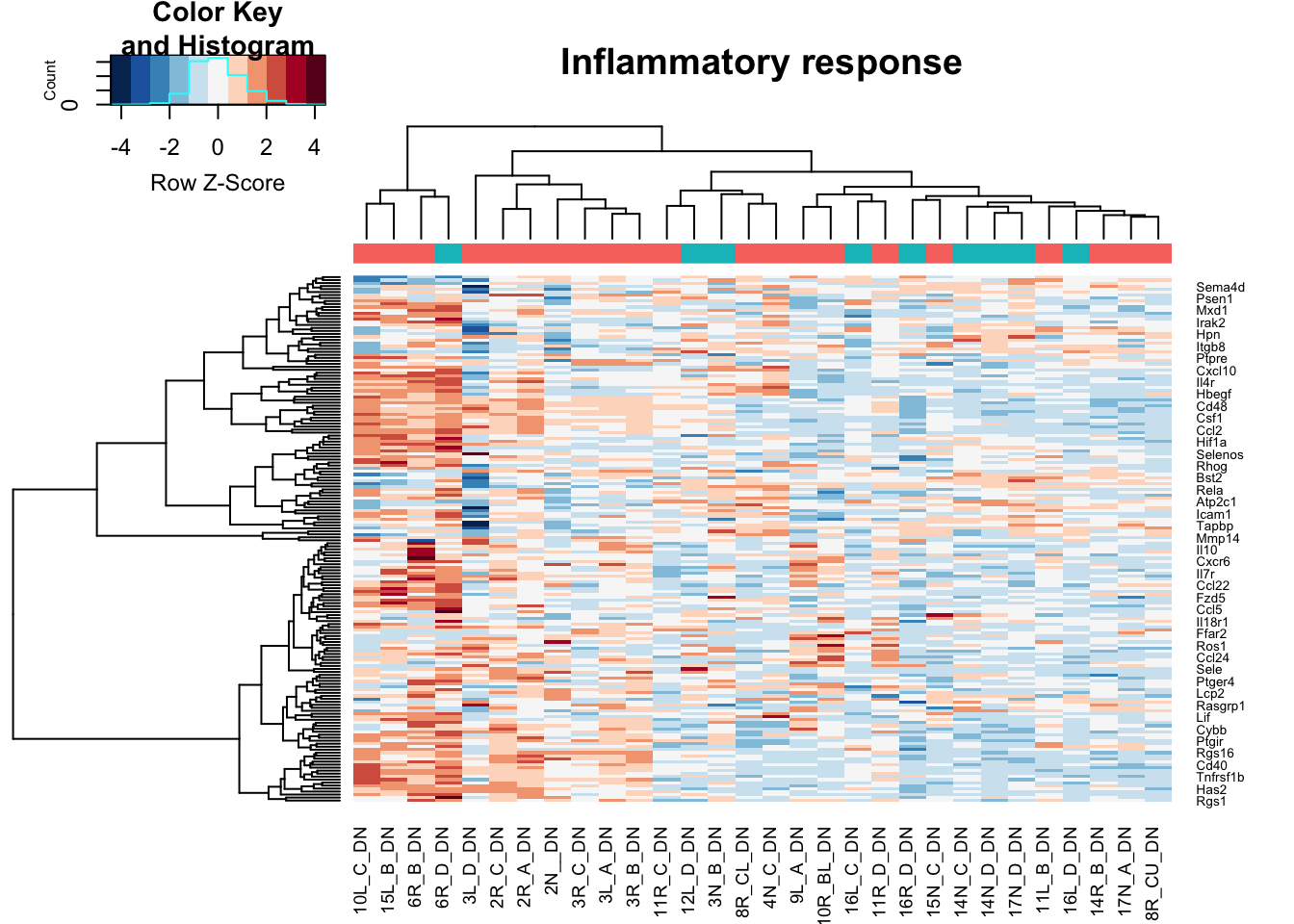
heatmap.2(assay(vstDN)[na.omit(match(RatGeneNamesCheckpoint, rownames(vstDN))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="checkpoint proteins")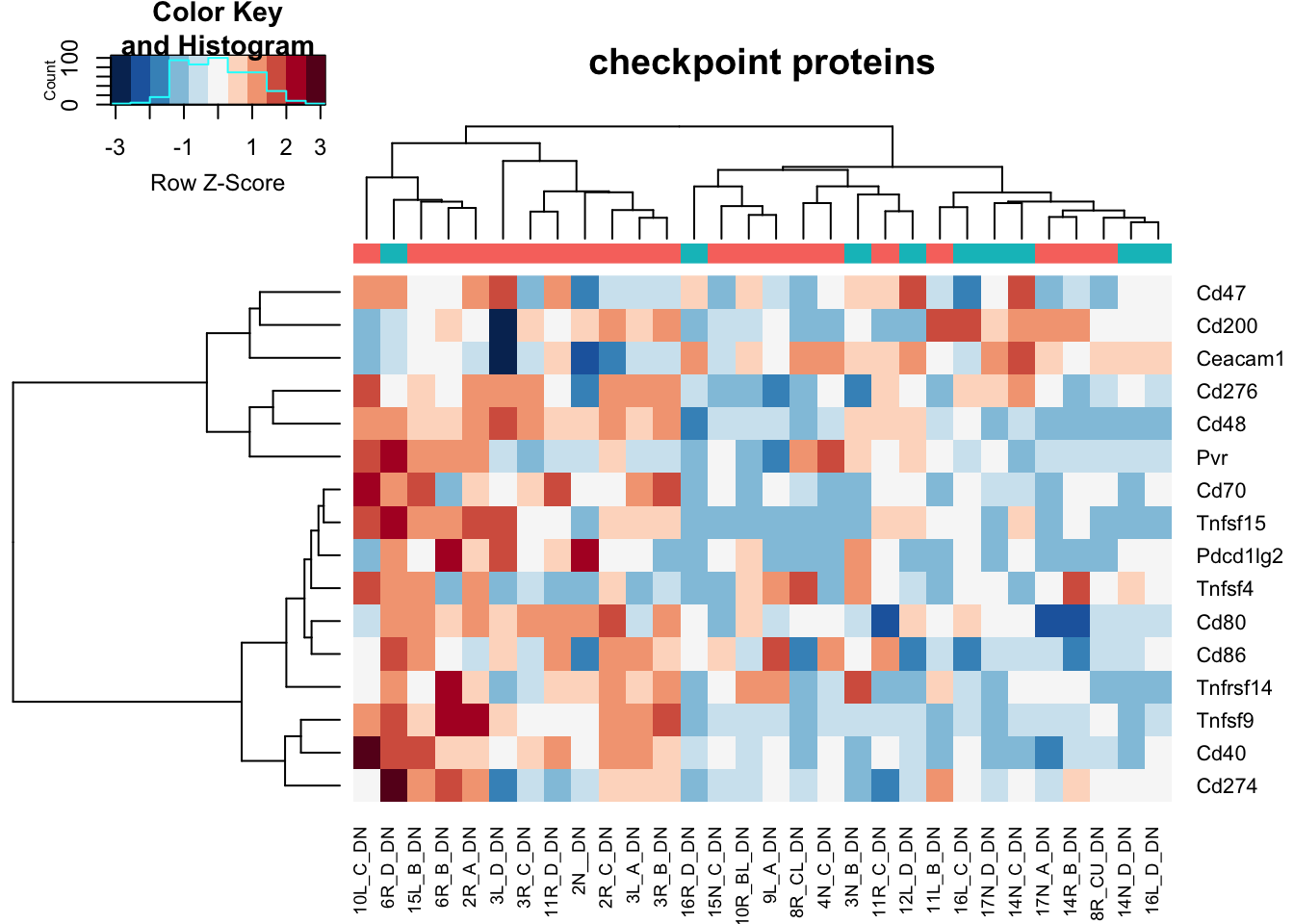
heatmap.2(assay(vstDN)[na.omit(match(MHCPres2Rat, rownames(vstDN))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC proteins")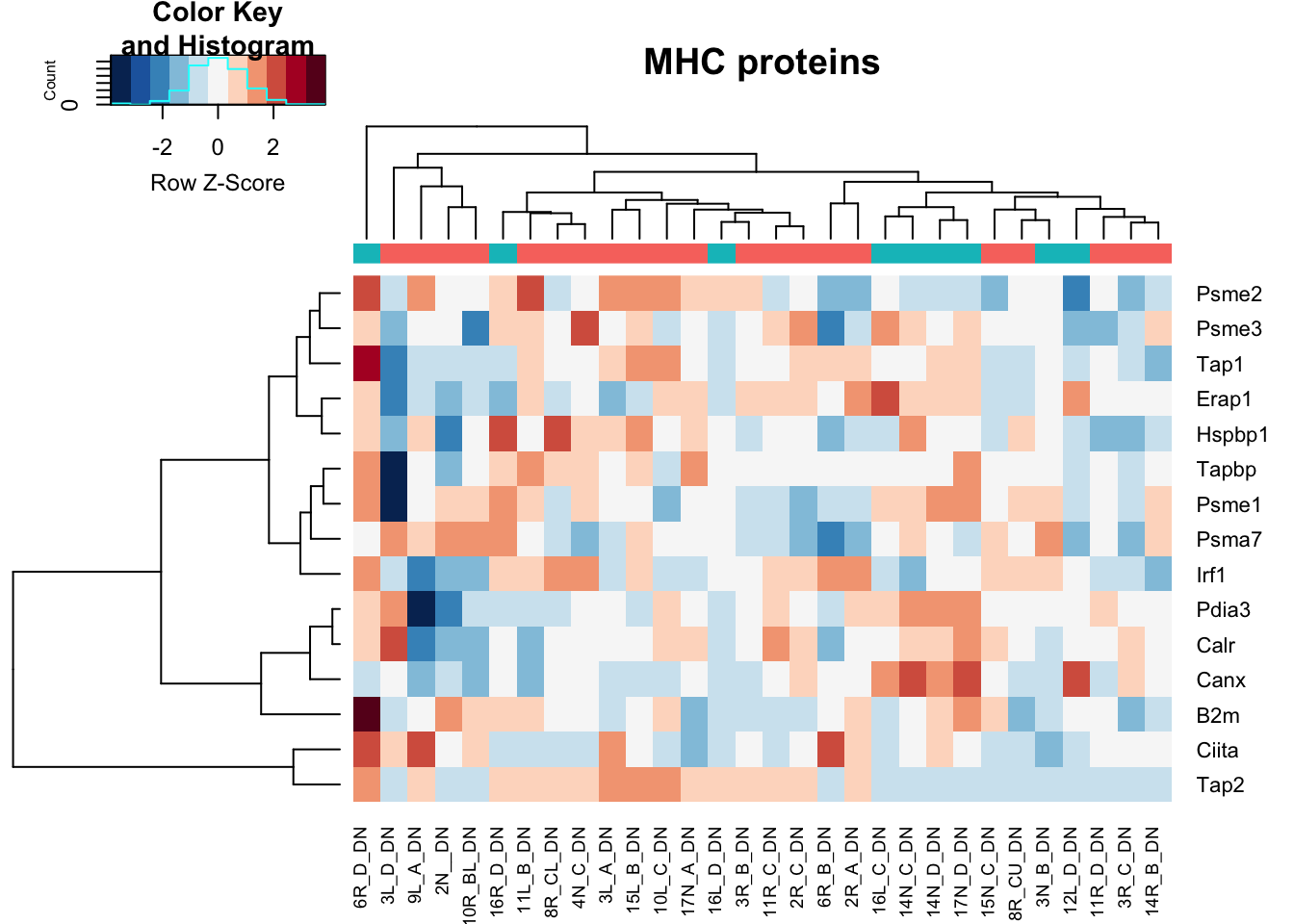
11.0.0.2 CD45 samples:
Note the distribution of growing/stable in CD is: 26, 8
ColsideColsC=hue_pal()(2)[CDdds$Growth]
heatmap.2(assay(vstCD)[na.omit(match(RatGeneNamesMHC, rownames(vstCD))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC expression")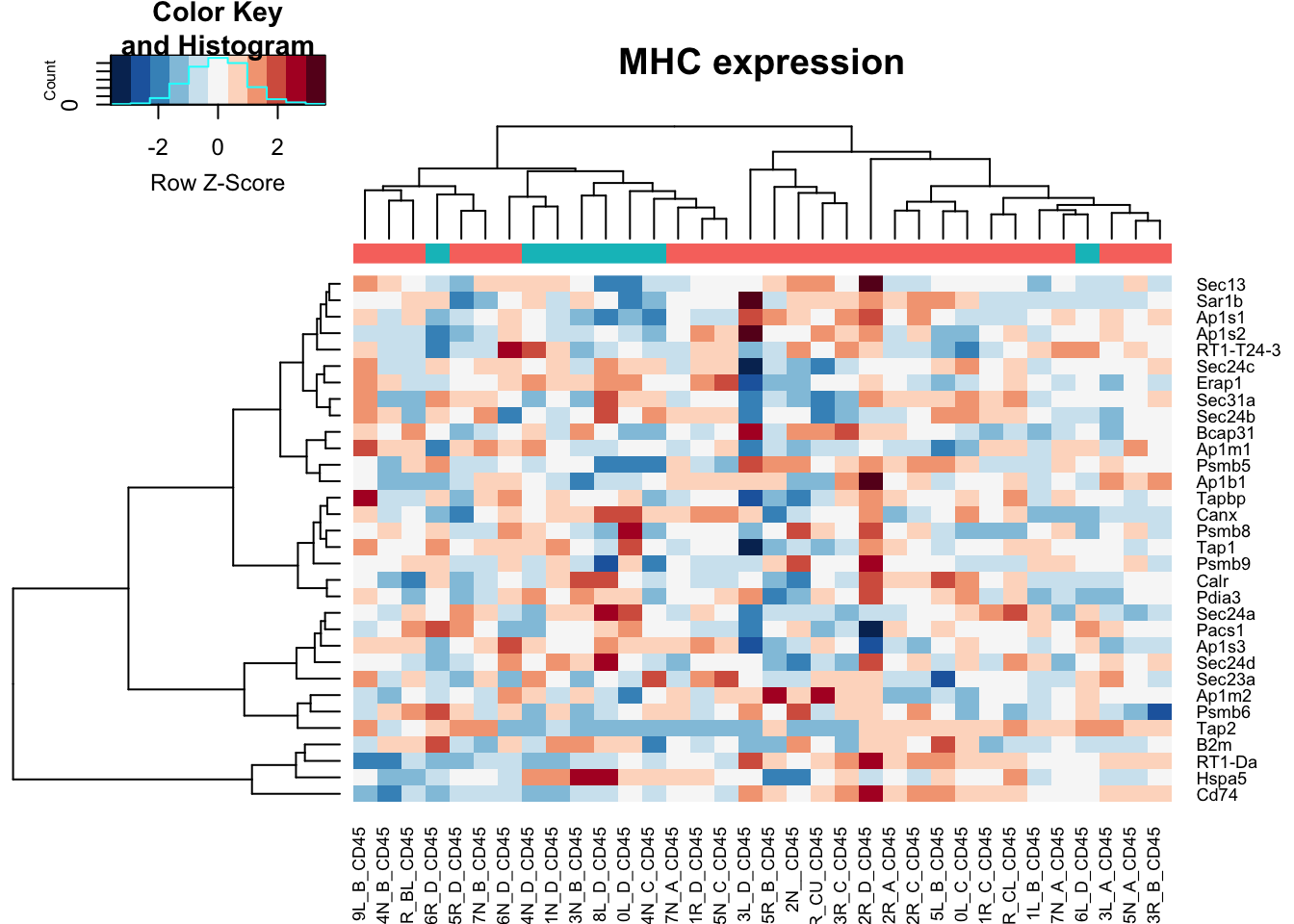
heatmap.2(assay(vstCD)[na.omit(match(RatGeneNamesKras, rownames(vstCD))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Kras expression")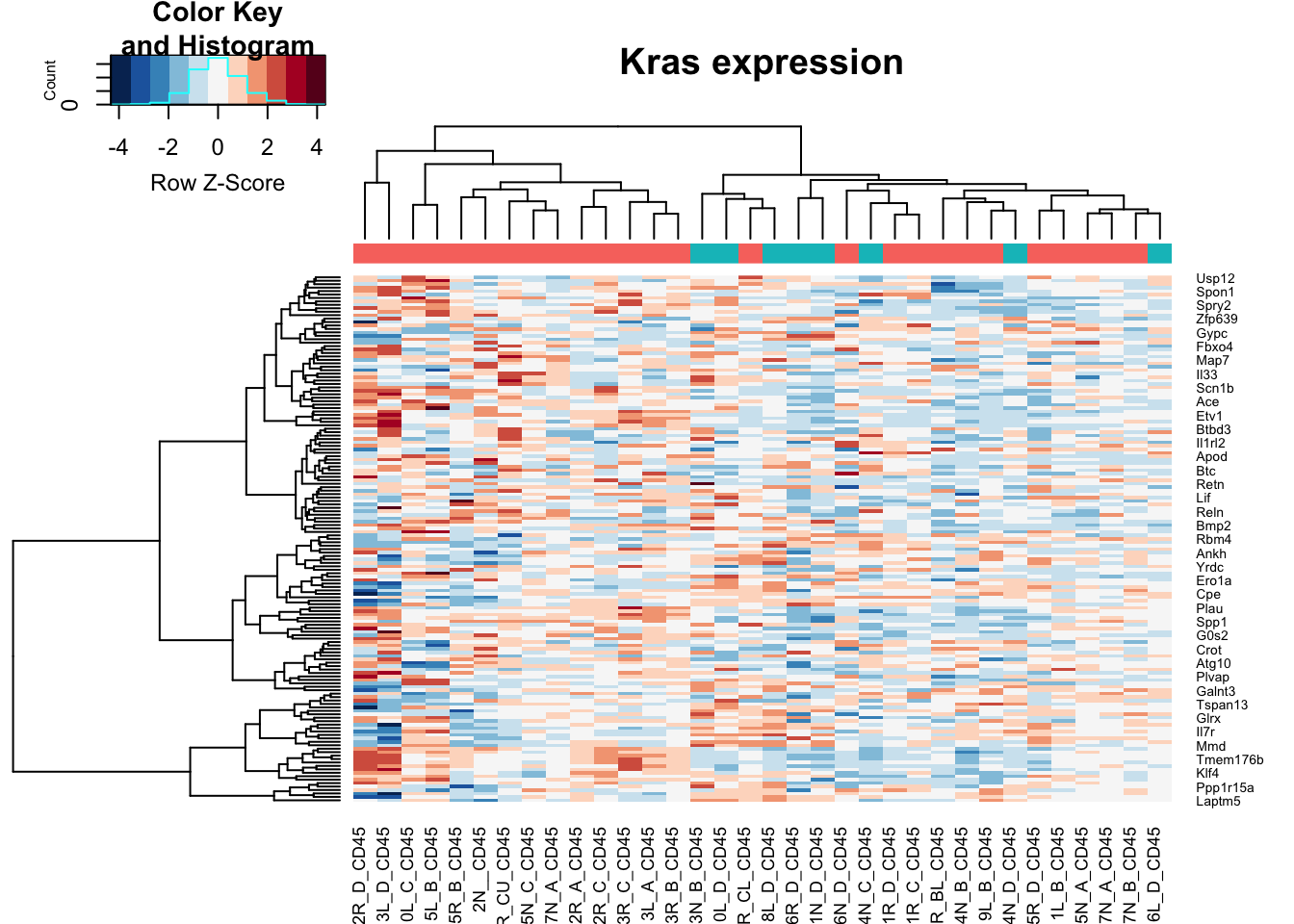
heatmap.2(assay(vstCD)[na.omit(match(RatGeneNamesInf, rownames(vstCD))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Inflammatory response")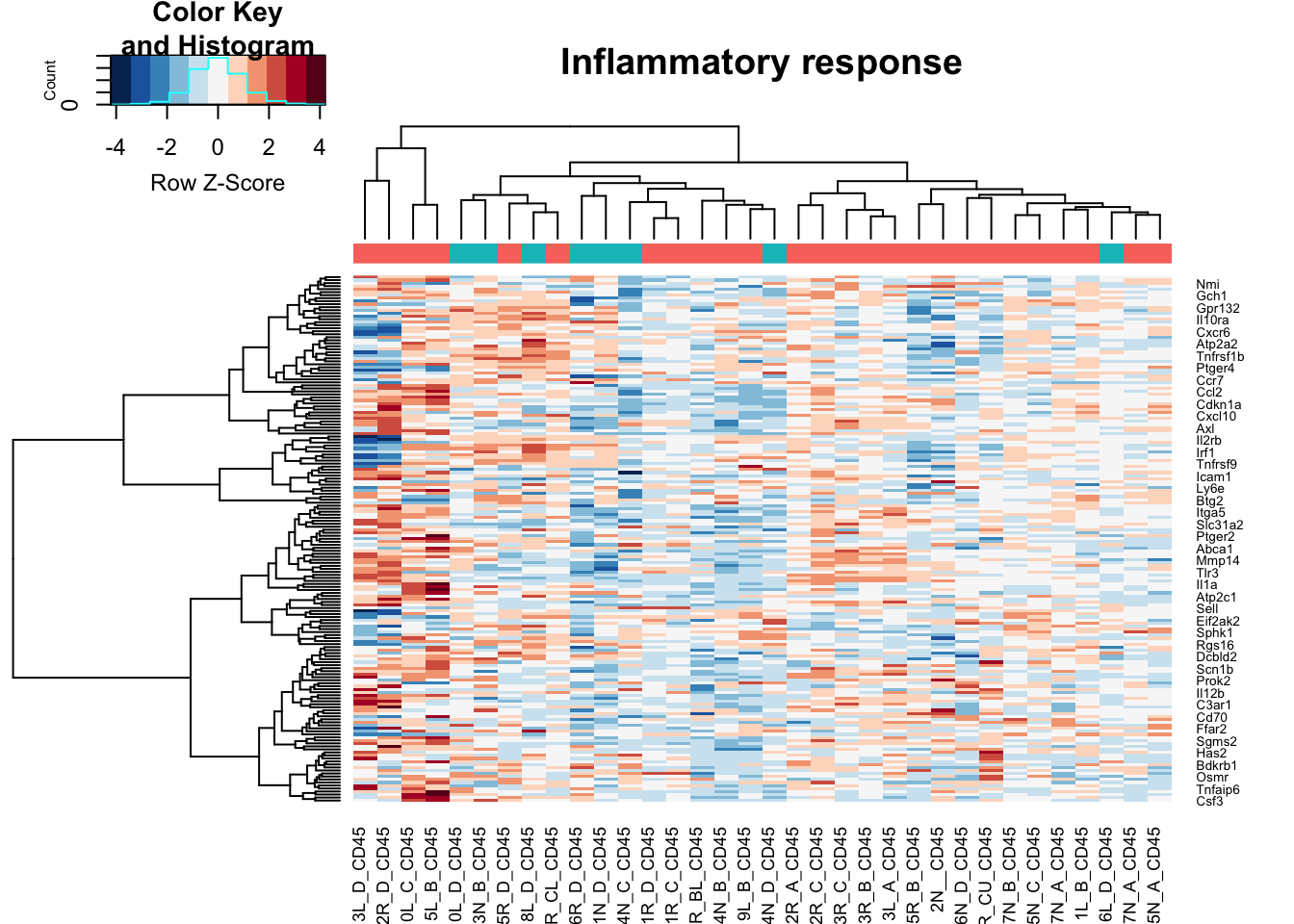
heatmap.2(assay(vstCD)[na.omit(match(RatGeneNamesCheckpoint, rownames(vstCD))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="checkpoint proteins")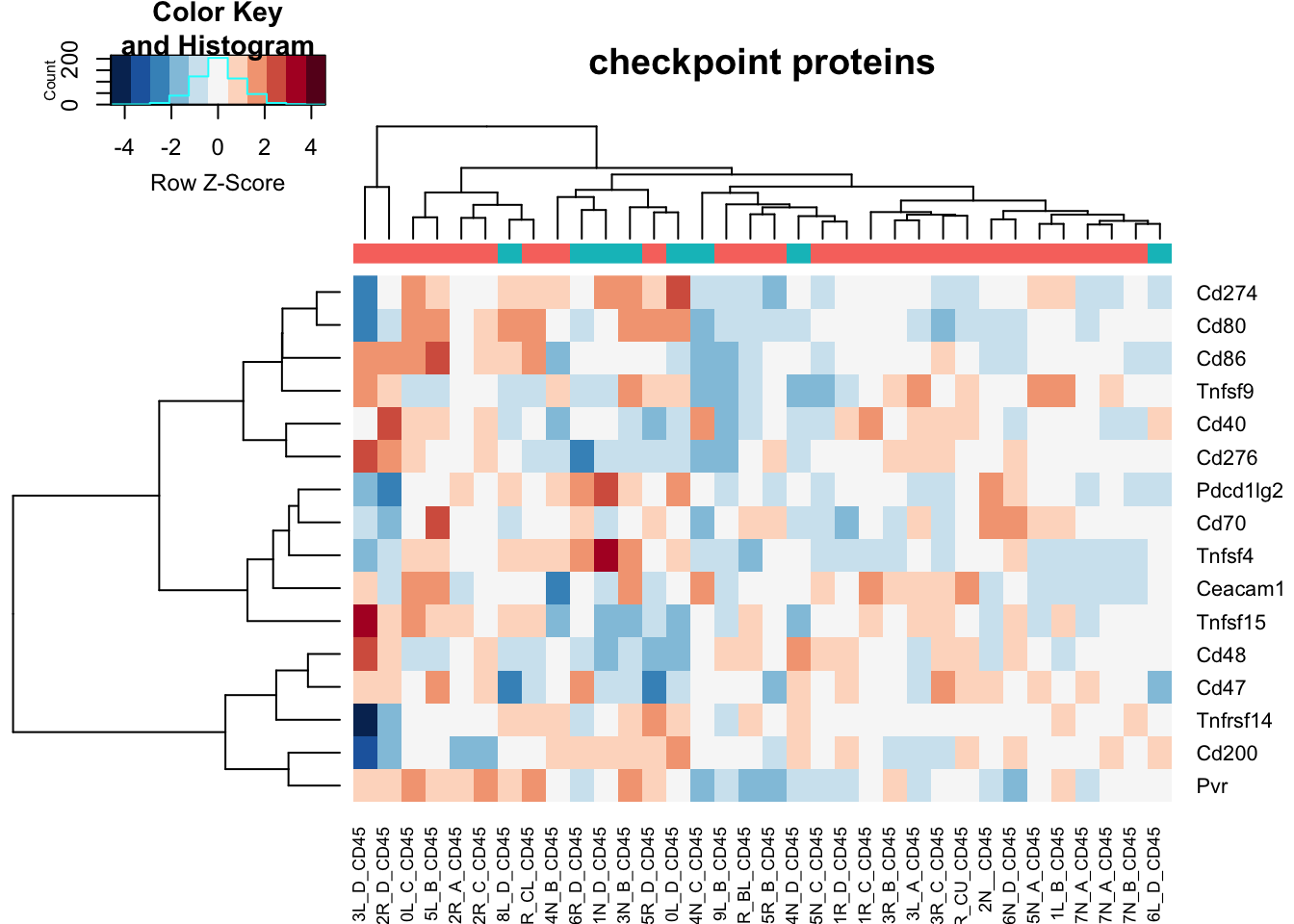
heatmap.2(assay(vstCD)[na.omit(match(MHCPres2Rat, rownames(vstCD))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC proteins")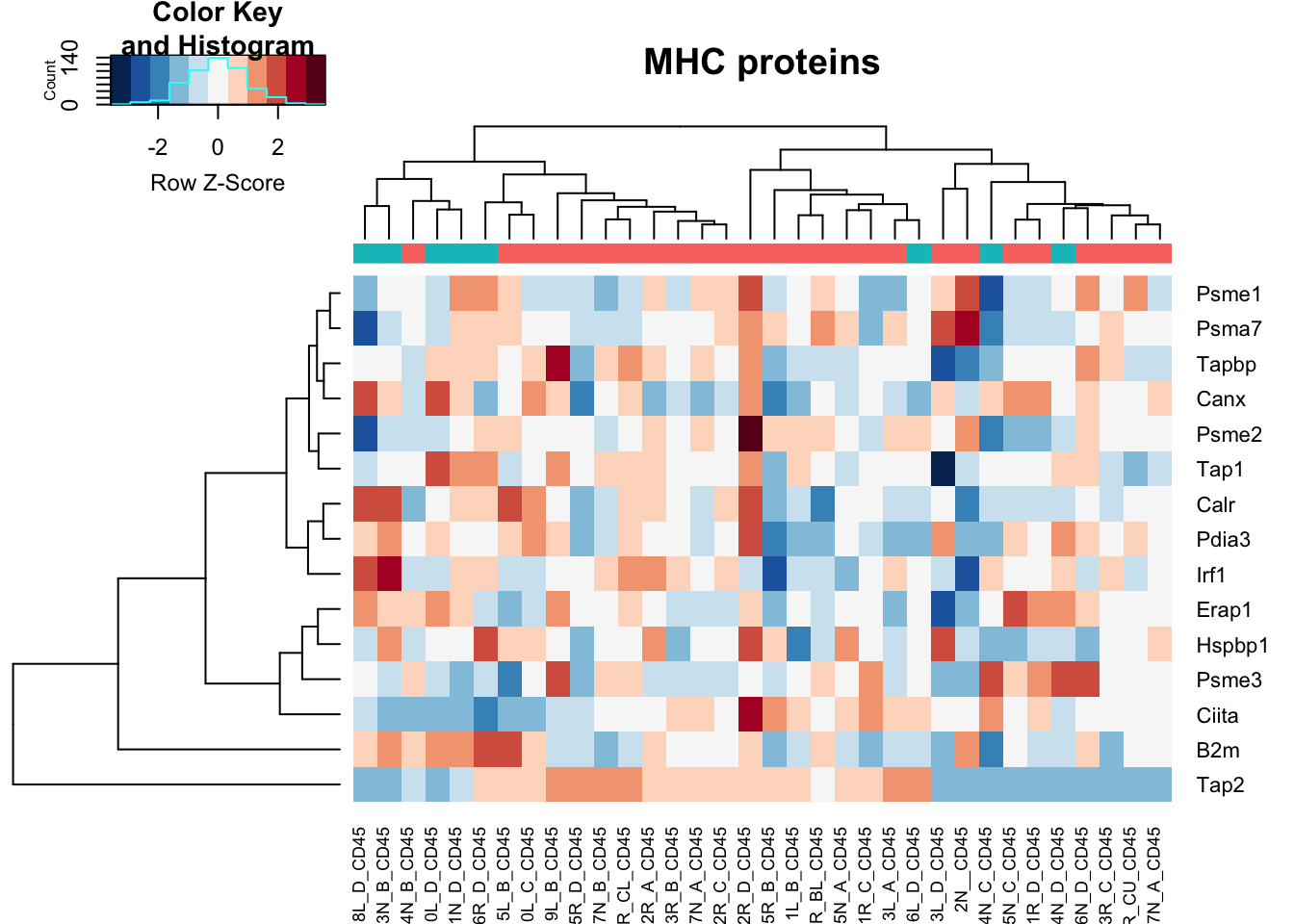
11.0.0.3 Epithelial samples:
ColsideColsC=hue_pal()(2)[Epdds$Growth]
heatmap.2(assay(vstEp)[na.omit(match(RatGeneNamesMHC, rownames(vstEp))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC expression")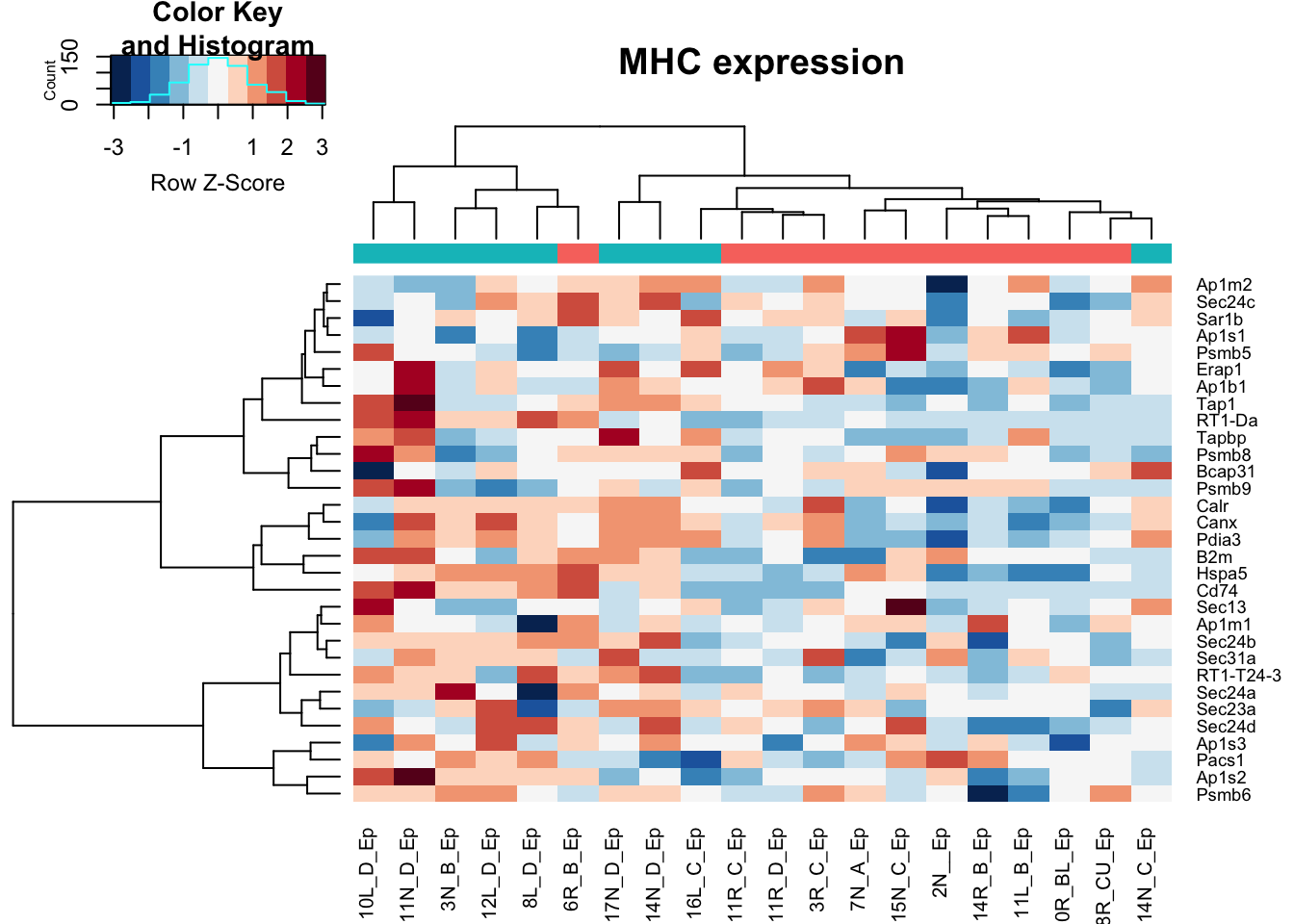
heatmap.2(assay(vstEp)[na.omit(match(RatGeneNamesKras, rownames(vstEp))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Kras expression")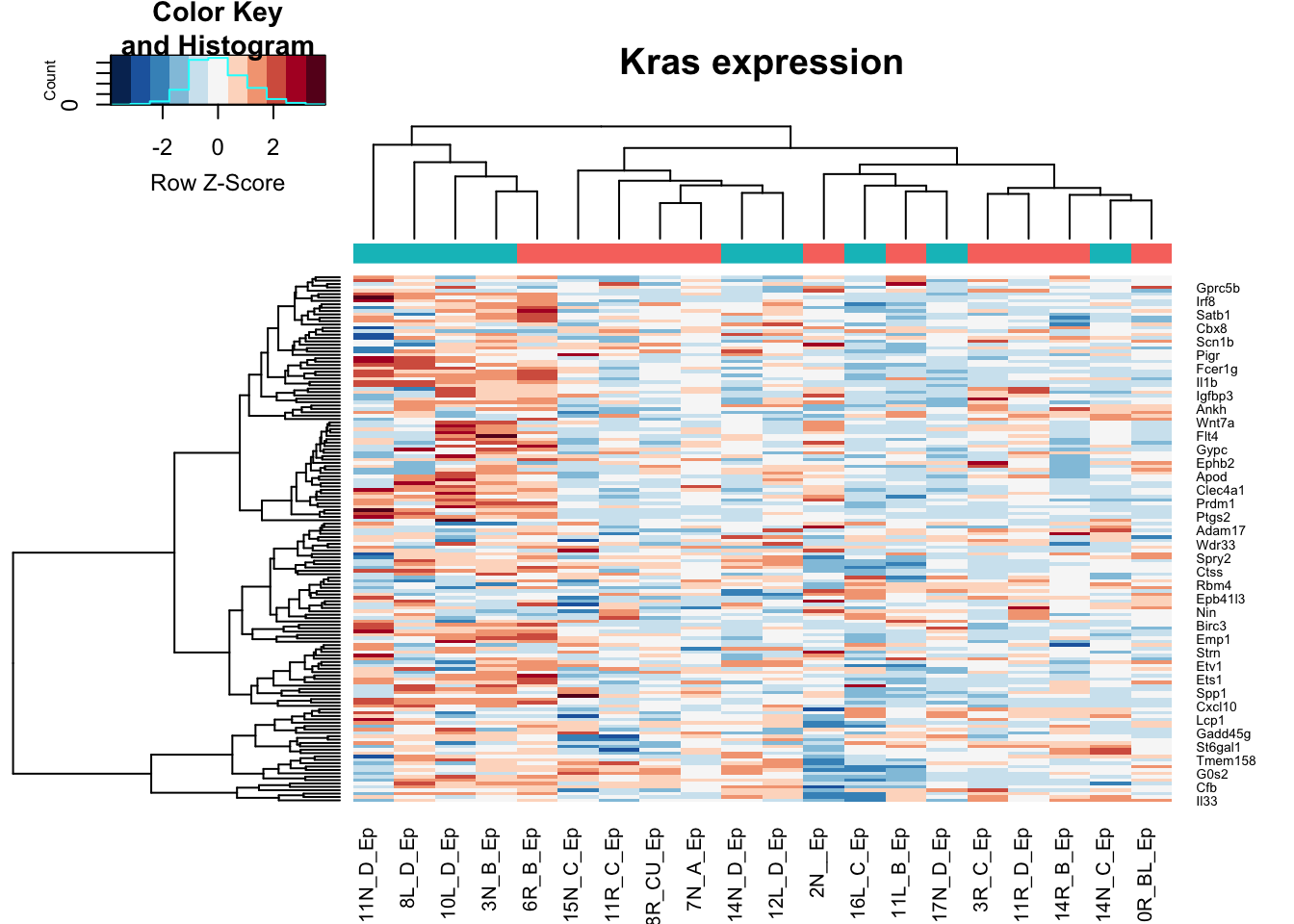
heatmap.2(assay(vstEp)[na.omit(match(RatGeneNamesInf, rownames(vstEp))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="Inflammatory response")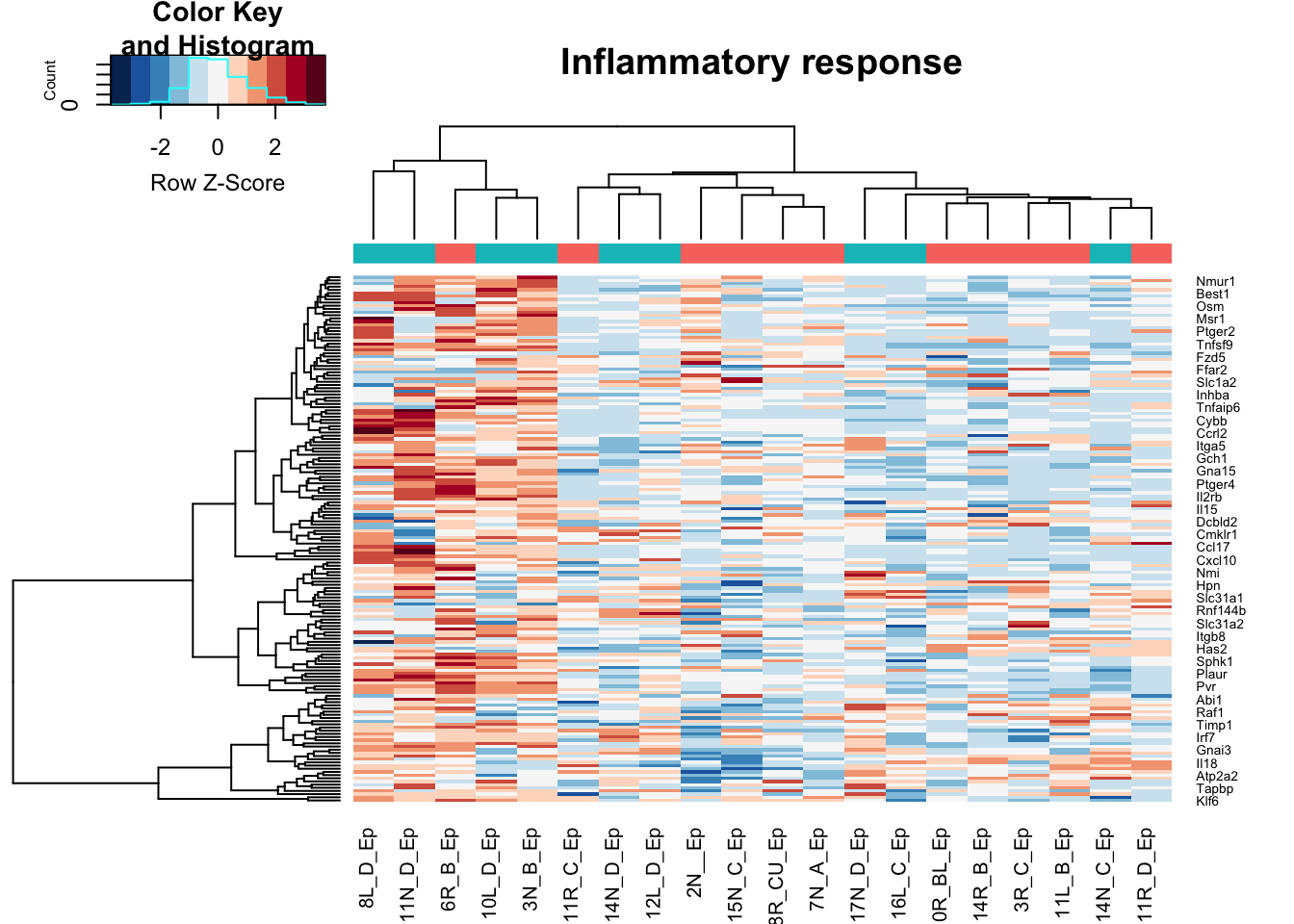
heatmap.2(assay(vstEp)[na.omit(match(RatGeneNamesCheckpoint, rownames(vstEp))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="checkpoint proteins")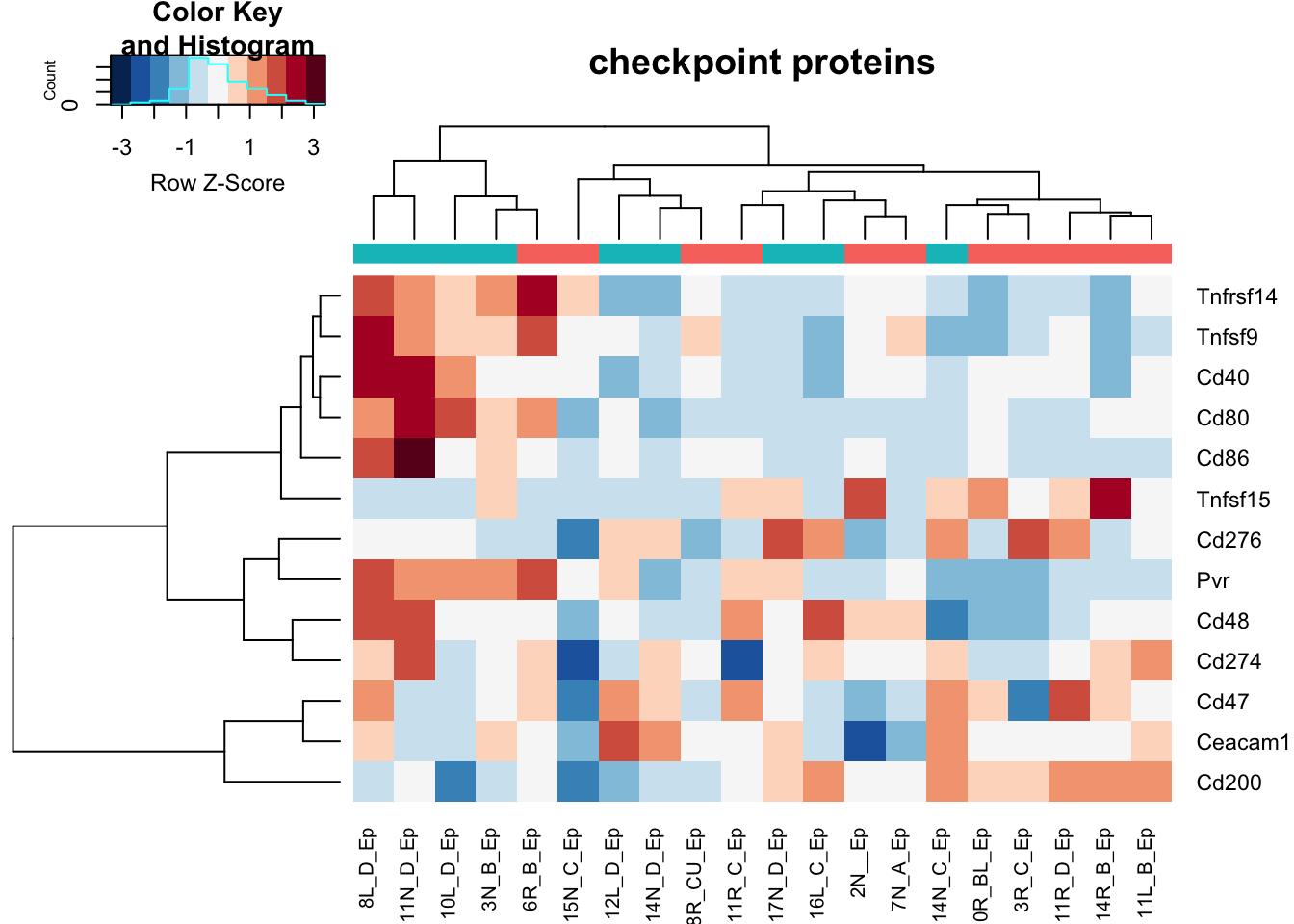
heatmap.2(assay(vstEp)[na.omit(match(MHCPres2Rat, rownames(vstEp))), ], col=RdBu[11:1], trace="none", scale="row", ColSideColors = ColsideColsC, main="MHC proteins")