Chapter 18 Signature analysis
In this section, we use ss gene-set enrichment analysis to investigate differences in
- Major signalling pathways
- MHC expression signatures
18.1 MHC signature analysis
Look if there is an association between MHC class I and class II and checkpoint proteins with growth in the different fractions. The summary appears:
- class I: association with stable in ep samples
- class II: low expression in DN stable sampels
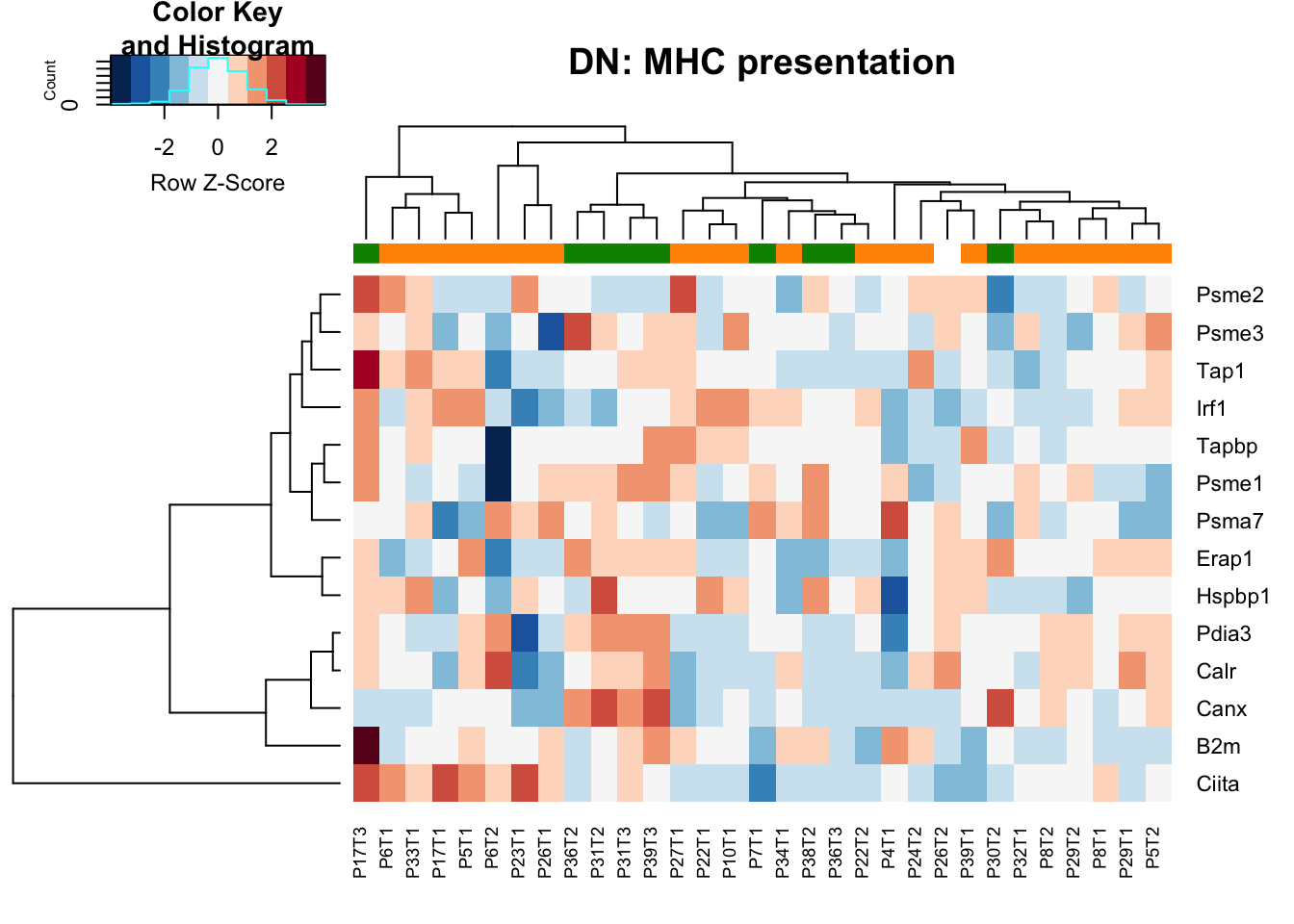
- MHC presentation: higher in stable samples Ep (and also the characterisation cohort in general), may also be the case in DN samples
18.1.1 MHC-I
classI <- c("RT1-A1", "RT1-A2", "RT1-A3", "RT1-Cl", "RT1-M2", "RT1-M3-1", "RT1-M4", "RT1-M5", "RT1-N1", "RT1-N2", "RT1-N3", "RT1-O1", "RT1-S2", "RT1-S3")
classII <- c("RT1-Ba", "RT1-Bb", "RT1-Da", "RT1-Db1", "RT1-Db2", "RT1-DMa", "RT1-DMb", "RT1-DOa", "RT1-DOb", "RT1-Ha")
#pdf(sprintf("rslt/signatureAnalysis/MHC_presentation_%s.pdf", Sys.Date()), height=6, width=7)
MHCclassSumm2=assay(vsd)[na.omit(match(classI, rownames(assay(vsd)))), ]
colnames(MHCclassSumm2)=infoTableFinal$TumorIDnew[match(colnames(MHCclassSumm2), rownames(infoTableFinal))]
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="CD45" & infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="CD45" & infoTableFinal$Cohort=="Progression")])], scale="row", main="CD: MHC class I", hclustfun = hclust.ave)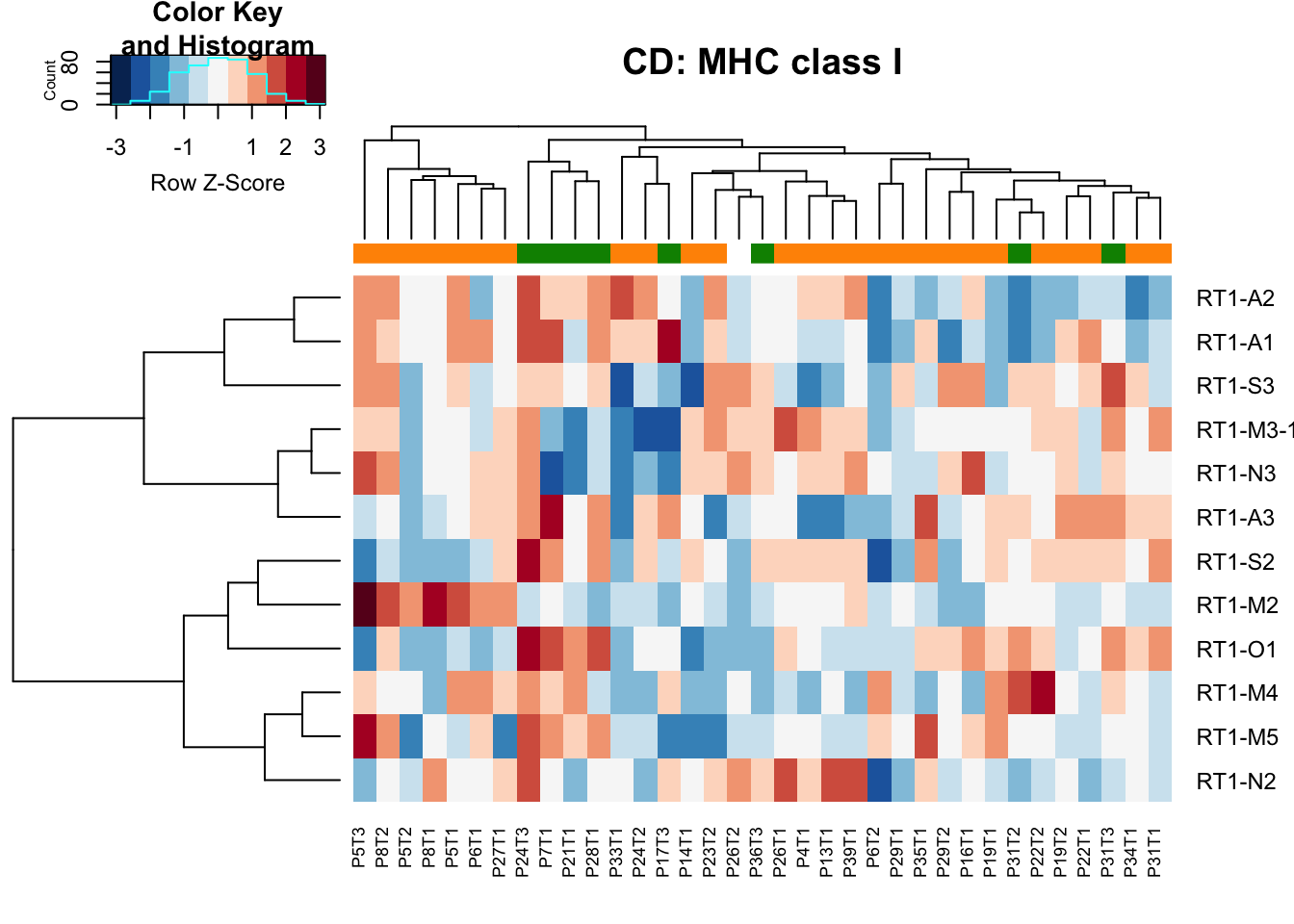
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="Ep" & infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="Ep" & infoTableFinal$Cohort=="Progression")])], scale="row", main="Ep: MHC class I", hclustfun = hclust.ave)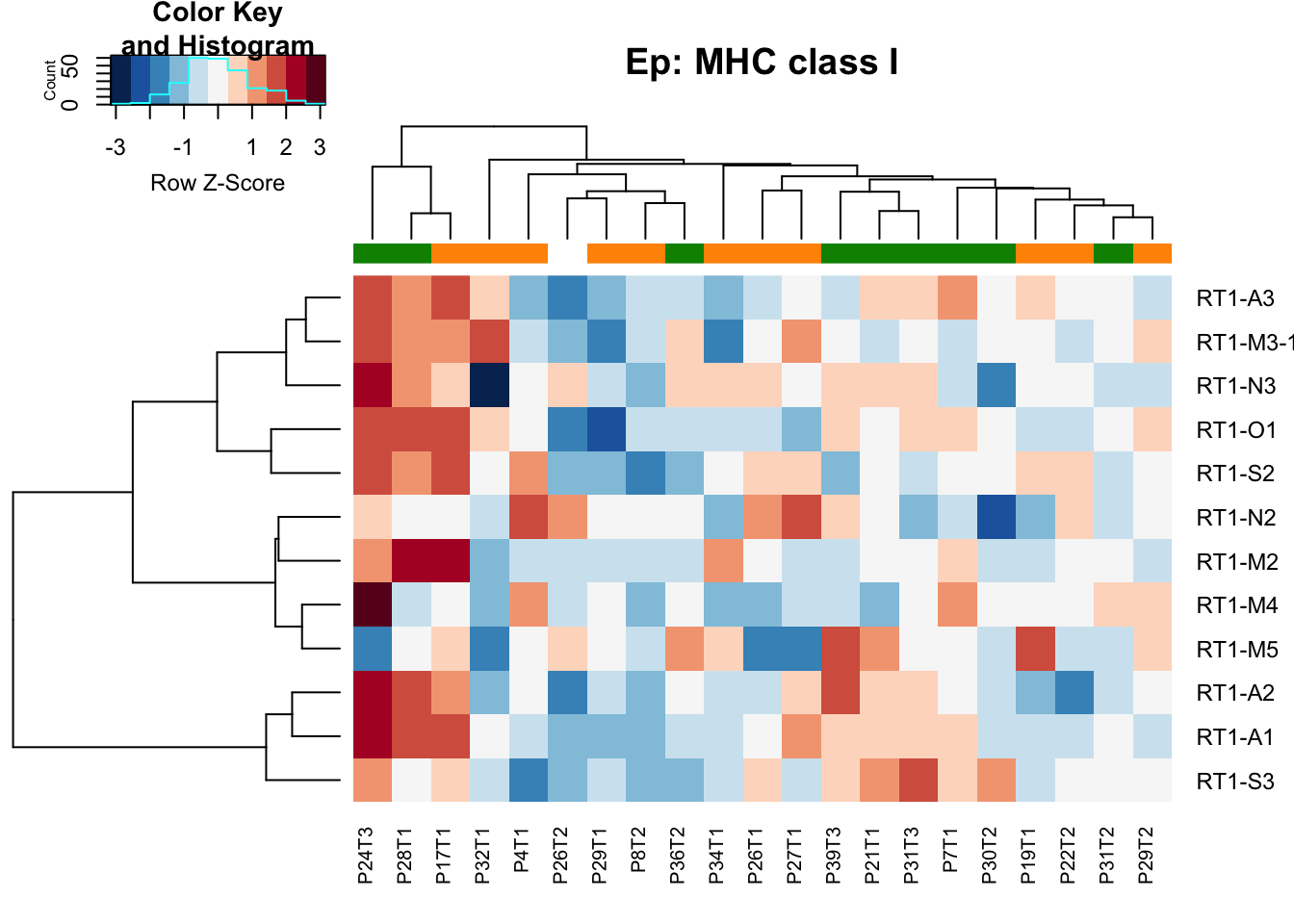
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="DN" & infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors =ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="DN" & infoTableFinal$Cohort=="Progression")])], scale="row", main="DN: MHC class I", hclustfun = hclust.ave)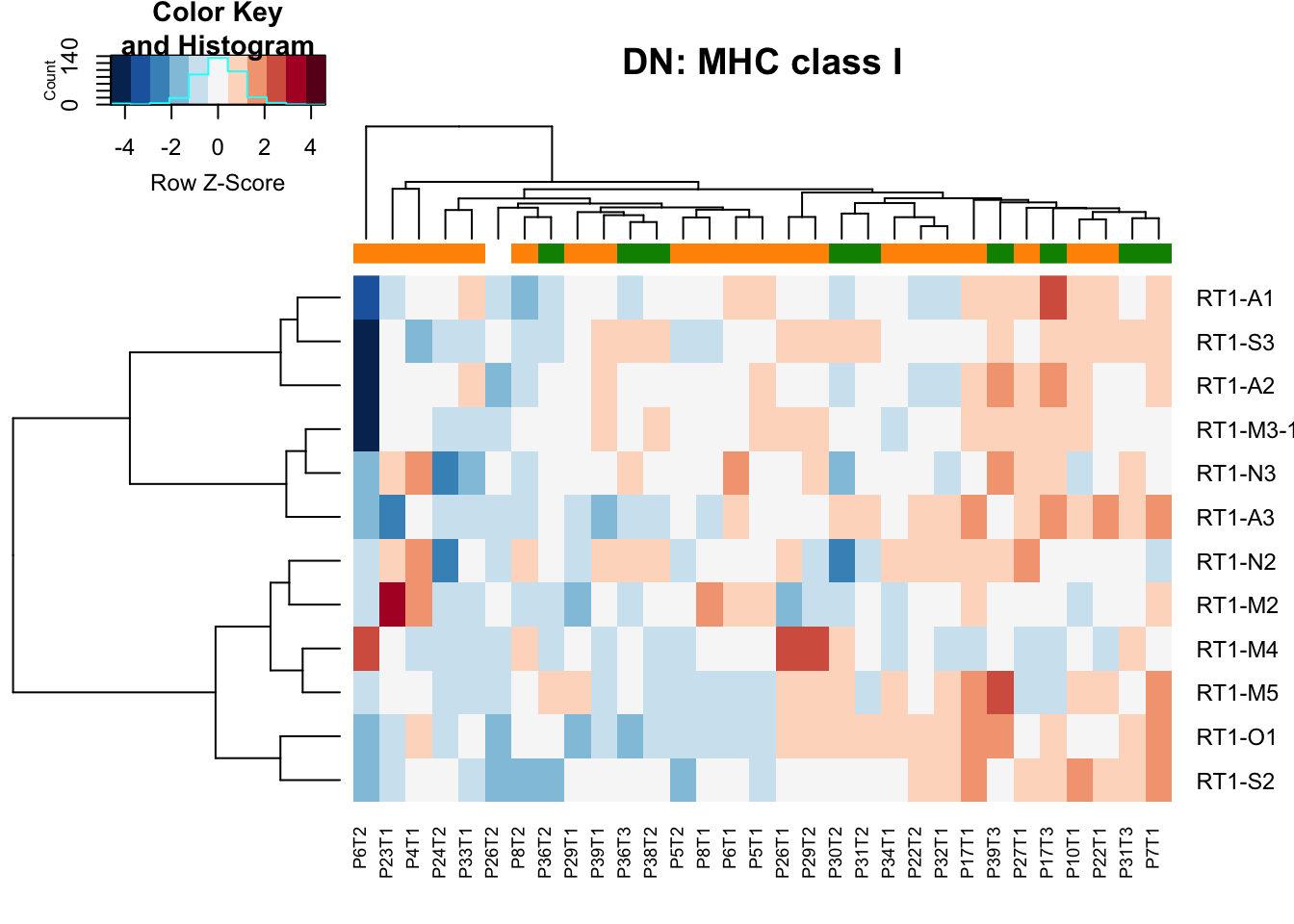
18.1.2 MHC-II
MHCclassSumm2=assay(vsd)[na.omit(match(classII, rownames(assay(vsd)))), ]
colnames(MHCclassSumm2)=infoTableFinal$TumorIDnew[match(colnames(MHCclassSumm2), rownames(infoTableFinal))]
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="CD45"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="CD45"& infoTableFinal$Cohort=="Progression")])], scale="row", main="CD: MHC class II")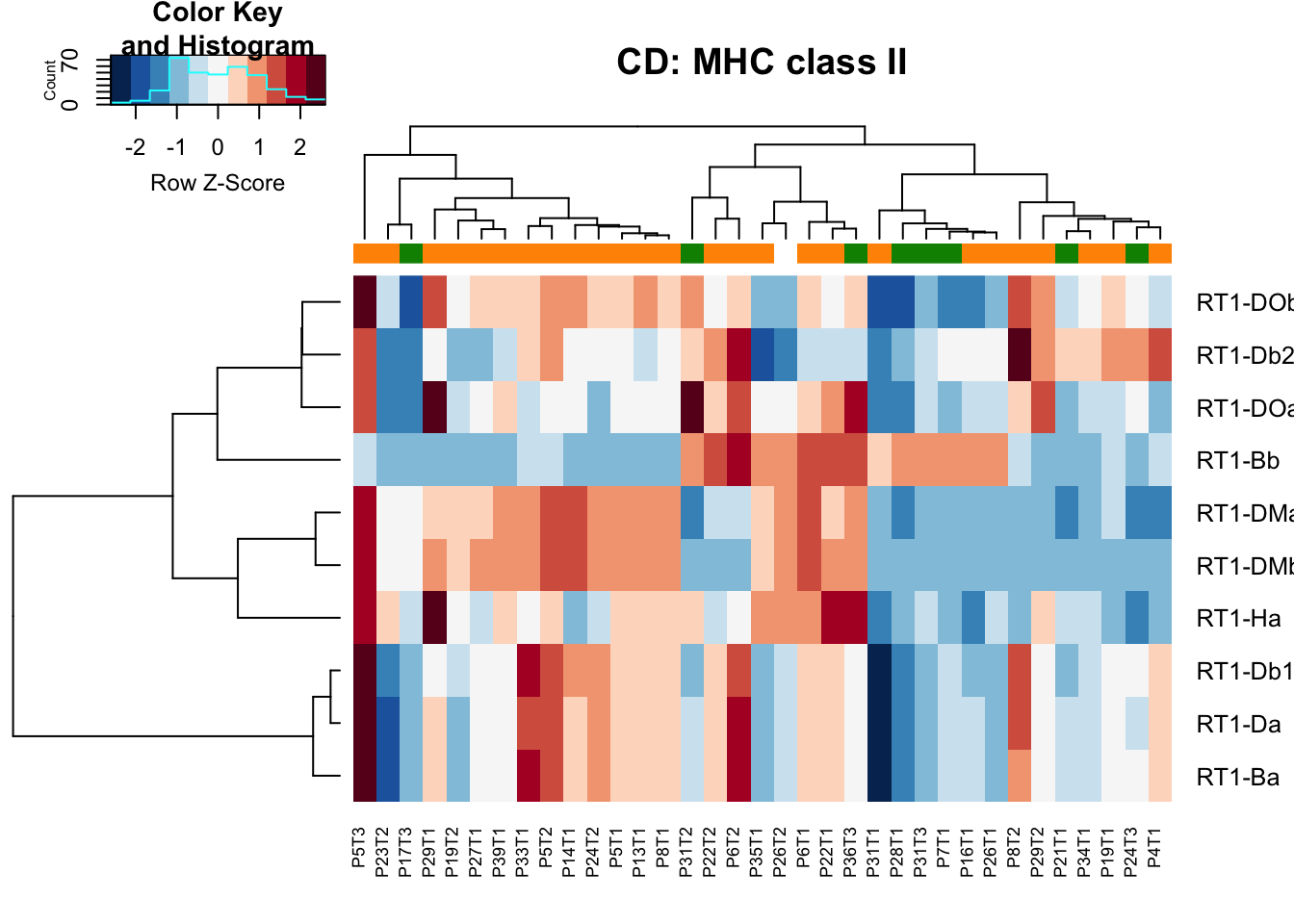
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="Ep"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="Ep"& infoTableFinal$Cohort=="Progression")])], scale="row", main="Ep: MHC class II")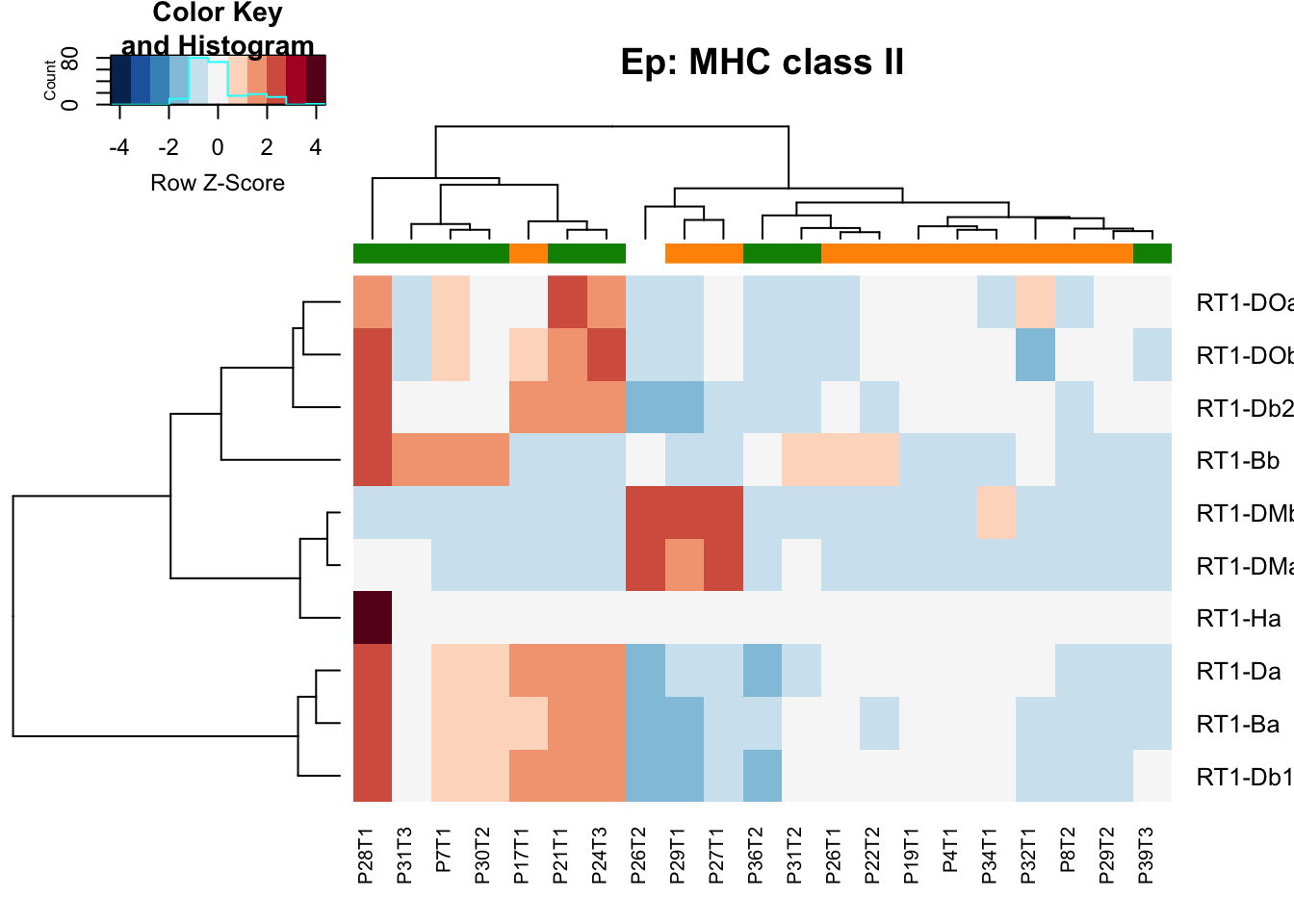
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="DN"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="DN"& infoTableFinal$Cohort=="Progression")])], scale="row", main="DN: MHC class II")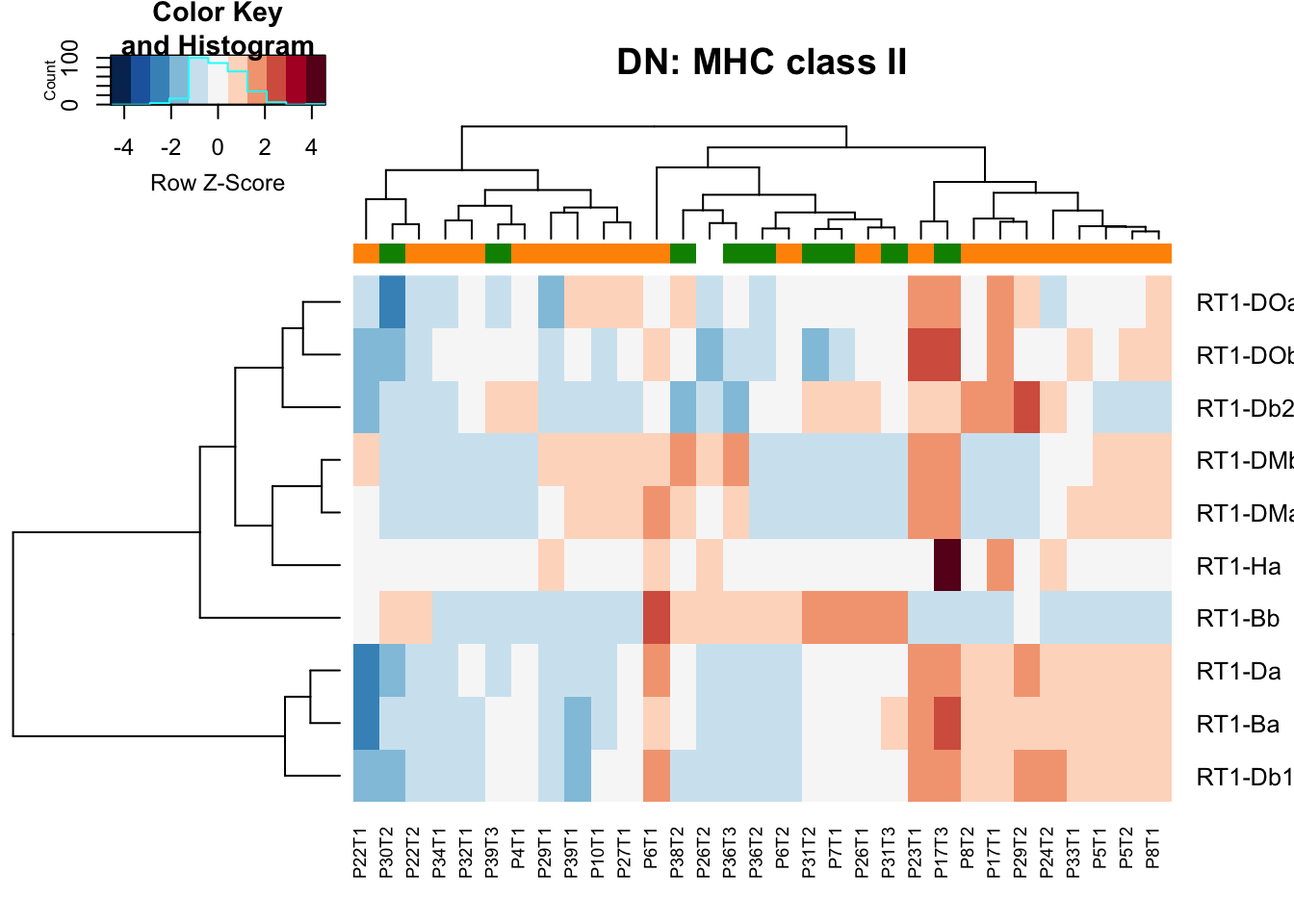
18.1.3 MHC presentation proteins
MHCclassSumm2=assay(vsd)[na.omit(match(MHCPres2Rat[-1], rownames(assay(vsd)))), ]
colnames(MHCclassSumm2)=infoTableFinal$TumorIDnew[match(colnames(MHCclassSumm2), rownames(infoTableFinal))]
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="CD45"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="CD45" & infoTableFinal$Cohort=="Progression")])], scale="row", main="CD45: MHC presentation")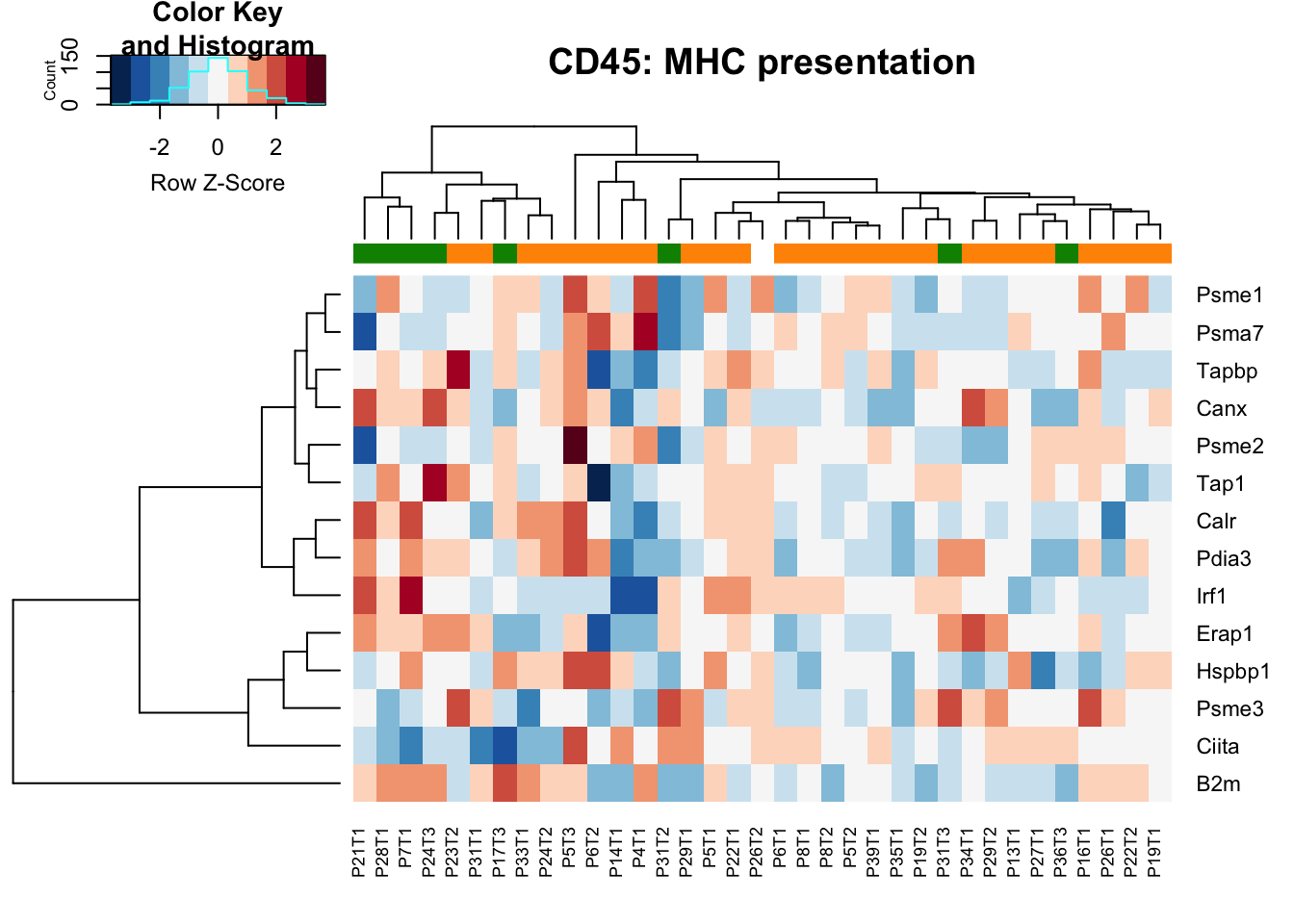
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="Ep"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="Ep"& infoTableFinal$Cohort=="Progression")])], scale="row", main="Ep: MHC presentation")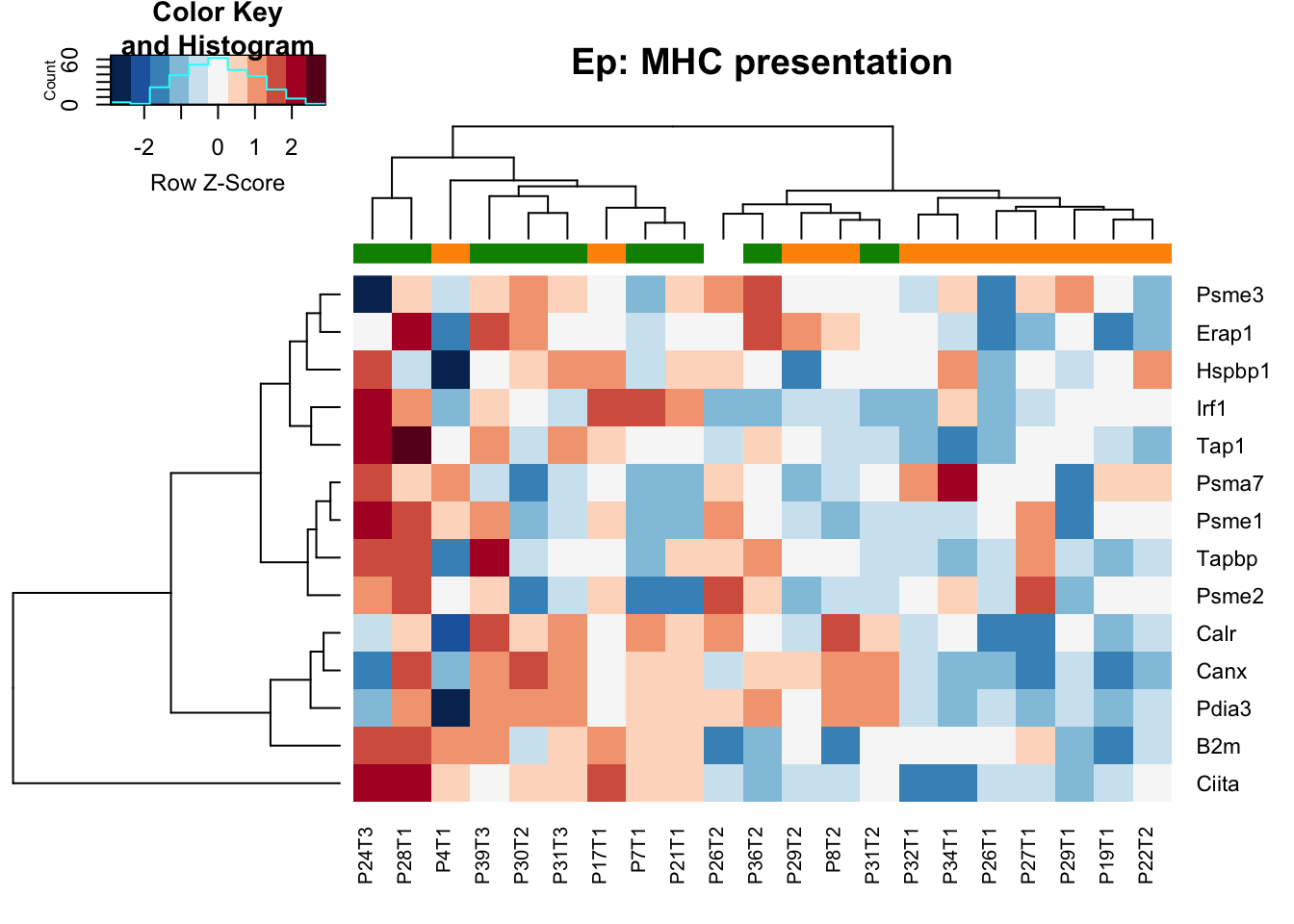
heatmap.2(MHCclassSumm2[ ,which(infoTableFinal$Fraction=="DN"& infoTableFinal$Cohort=="Progression")], col=RdBu[11:1], trace="none", ColSideColors = ColSizeb[factor(infoTableFinal$Growth[which(infoTableFinal$Fraction=="DN"& infoTableFinal$Cohort=="Progression")])], scale="row", main="DN: MHC presentation")